Software
Sparse Generalized Correlation Analysis
Sparse GCA
\[\max_L \mathrm{Tr}(L^\top\Sigma L), \quad s.t.\quad L^\top\Sigma_0 L=I_r, \quad ||L||_{2, 0} \leq s\]Generalized correlation analysis (GCA) is concerned with uncovering linear relationships across multiple datasets. It generalizes canonical correlation analysis that is designed for two datasets. We study sparse GCA when there are potentially multiple generalized correlation tuples in data and the loading matrix has a small number of nonzero rows. It includes sparse CCA and sparse PCA of correlation matrices as special cases. We first formulate sparse GCA as generalized eigenvalue problems at both population and sample levels via a careful choice of normalization constraints. Based on a Lagrangian form of the sample optimization problem, we propose a thresholded gradient descent algorithm for estimating GCA loading vectors and matrices in high dimensions. We derive tight estimation error bounds for estimators generated by the algorithm with proper initialization. We also demonstrate the prowess of the algorithm on a number of synthetic datasets. R and MATLAB code can be found here.
CellLENS
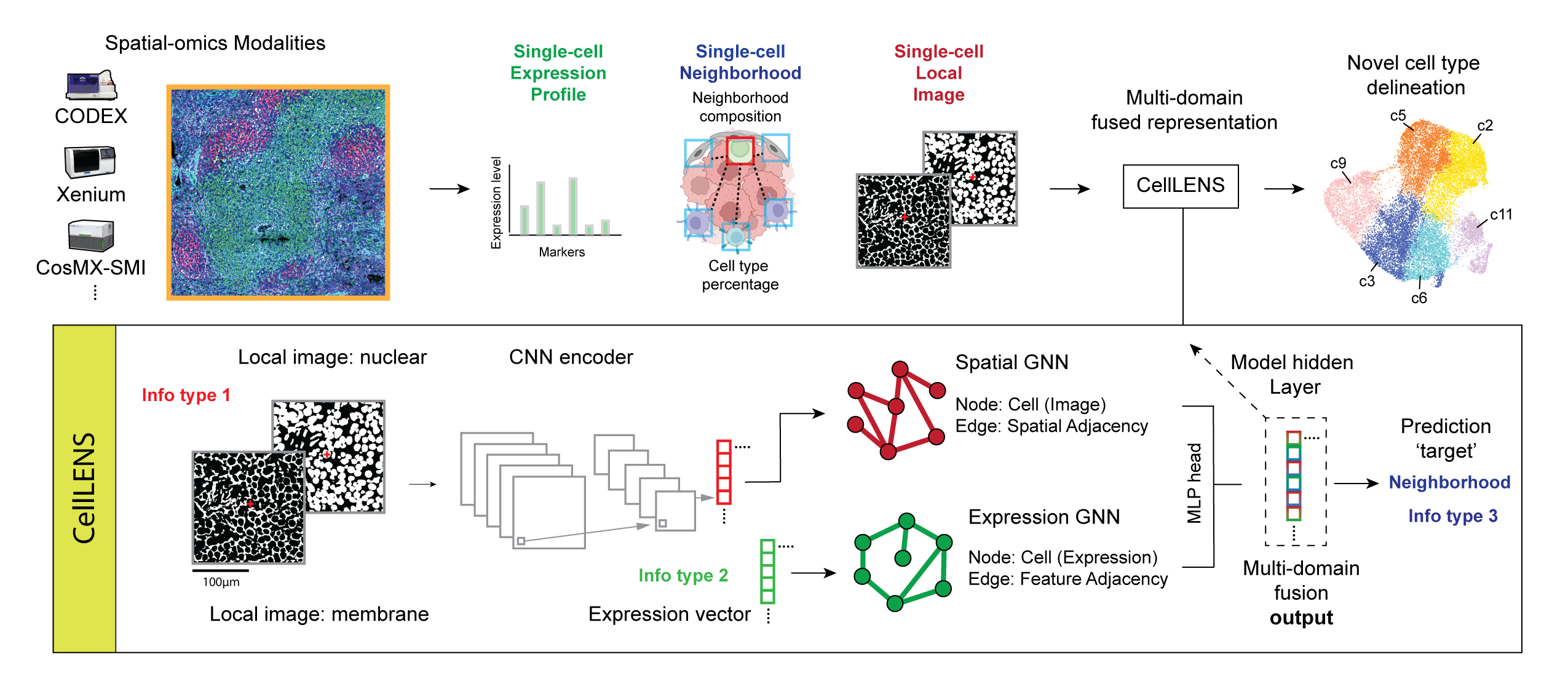
Cell population identification is a crucial and necessary step in almost all single-cell studies. The current approach for clustering and annotating cells in spatial-omics datasets mirrors methods originally established for dissociated single-cell methods. In this process, spatial-image level information is initially condensed into a non-spatial expression profile, followed by de novo cell type identification pipelines. However, this limited information represents a missed opportunity to uncover cell populations functionally stratified by their spatial components: a key motivation for employing spatial-omics technologies in the first place. To address this gap, we introduce Cell Local Environment Neighborhood Scan (CellLENS), a computational method that learns a single-cell representation embedding by integrating cross-domain information from tissue samples. Through the analysis of datasets spanning spatial proteomic and spatial transcriptomic modalities, and across different tissue types and disease settings, we demonstrate CellLENS’s capability to elucidate biologically relevant cell populations that were previously elusive due to the relinquished tissue morphological information from images.
Official implementation of CellLENS package can be found here.




